The ability to engineer the mouse genome has proven useful for a variety of applications in research, medicine and biotechnology. Transgenic mice have become powerful reagents for modeling genetic disorders, understanding embryonic development and evaluating therapeutics. These mice and the cell lines derived from them have also accelerated basic research by allowing scientists to assign functions to genes, dissect genetic pathways, and manipulate the cellular or biochemical properties of proteins.
Gene targeting
Homologous recombination in embryonic stem cells is now a routine method for modifying the mouse genome at a specific locus. The technique was first developed for site-directed mutagenesis in yeast, and has been successfully adapted for mammalian cells (7). Any deletion, point mutation, inversion or translocation can now be modeled in mice. This is accomplished by generating a piece of DNA that is identical to the locus of interest – except for the alteration and a drug resistance marker- and this engineered piece is swapped in to replace the original piece of DNA. The DNA construct to be introduced into the genome of the ES cells should contain the mutation with several kilobases of DNA that are homologous to the mouse genome flanking the mutation. These flanking sections are where the recombination must occur. Homologous recombination in ES cells is a very rare event (less than 0.01%), so the vector must contain genes conferring drug resistance or sensitivity so researchers can enrich their population for cells that have taken up the DNA. Even with this selection, most of the surviving ES cells have integrated the new piece of DNA at a random locus rather than recombining it at the correct locus of interest. Due to this, ES cells showing resistance to the selective agent must also be screened by Southern blot or by PCR to discover which clones have been correctly targeted.
Knockin Mice
To avoid the problems of a standard transgenic, many researchers now rely on knockin mice to study the exogenous expression of a protein. A knockin mouse is generated by targeted insertion of the transgene at a selected locus. The insert is flanked by DNA from a non-critical locus, and homologous recombination allows the transgene to be targeted to that specific, non-critical integration site. (See Figure 1) In this way, a researcher has complete control of the genetic environment surrounding the overexpression cassette and it is likely that the DNA did not incorporate itself into multiple locations. Site-specific knockins result in a more consistent level of expression of the transgene from generation to generation because it is known that the overexpression cassette is present as a single copy. Also, because a targeted transgene is not interfering with a critical locus, the researcher can be more certain that any resulting phenotype is due to the exogenous expression of the protein. Although the generation of a knockin mouse does avoid many of the problems of a traditional transgenic mouse, this procedure requires more time to assemble the vector and to identify ES cells that have undergone homologous recombination.
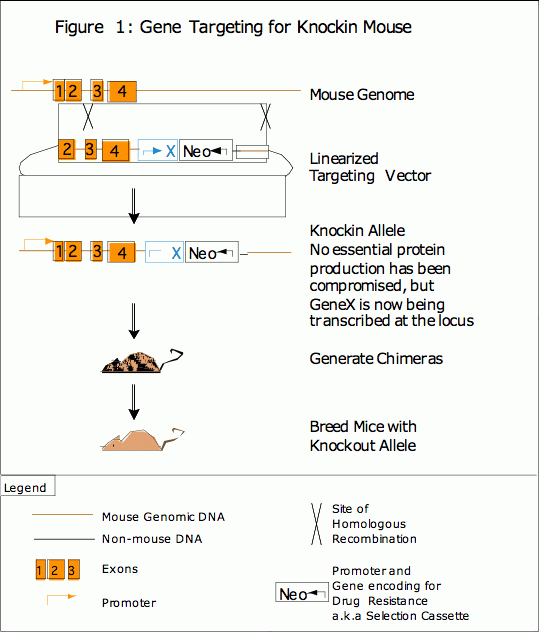
While traditional transgenic and knockin mice are generated to express a protein, much information can be learned from the elimination of a gene or the deletion of a functional domain of the protein. This can be achieved through random mutation using chemical mutagenesis or a gene trap approach, or through gene targeting to generate a knockout mouse. Homologous recombination allows a researcher to completely remove one or more exons from a gene, (see Figure 2) which results in the production of a mutated or truncated protein or, more often, no protein at all. (See Figure 2).
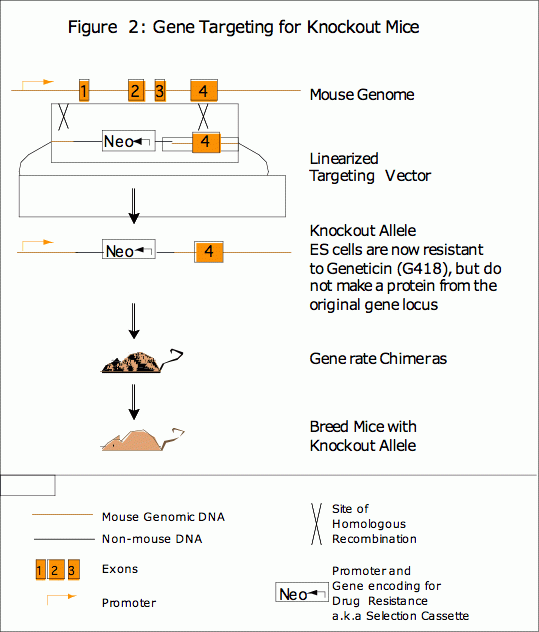
The phenotypes of knockout mice can be very complex because all tissues of the mouse may be affected, though it is not uncommon for a knockout mouse to display embryonic lethality or to show no phenotype at all. The process and time line for making a knockout mouse with the KI Preclinical Modeling Core Facility has been outlined as a Gene Targeting Timeline in the Services section.
Conditional gene modification
Many genes that participate in interesting genetic pathways are essential for either mouse development, viability or fertility. Therefore, a traditional knockout of the gene can never lead to the establishment of a knockout mouse strain for analysis. Conditional gene modification using Cre-lox and Flp-frt technology allows the gene of interest to be knocked-out in only a subset of tissues or only at a particular time, circumventing lethality. Because gene targeting can be controlled both spatially and temporally, the function of a given gene can be studied in the desired cell types and at a specific time point. This genetic dissection allows researchers to define gene function in development, physiology or behavior.
Cre recombinase, a site-specific integrase isolated from the P1 bacteriophage, catalyzes recombination between two of its consensus DNA recognition sites (8). These loxP sites are 34 base pairs in length, consisting of two 13bp palendromic sequences that flank a central sequence of 8bp which determines the directionality of the loxP site. Two loxP sites are most often placed in a trans orientation on either side of an essential, functional part of a gene so that recombination removes that functionality and knocks-out the gene. (See Figure 3) LoxP sites can also be placed in a cis orientation to invert the intervening sequence. LoxP sites placed on different chromosomes can be used to generate targeted translocations, though this recombination event occurs at a relatively low frequency compared to the highly-efficient intra-gene recombination.
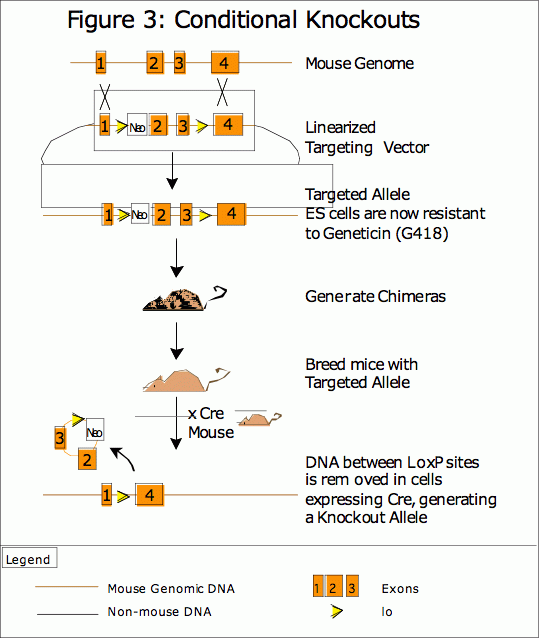
LoxP sites do not recombine in the absence of Cre recombinase, so regulation of the expression of Cre recombinase also regulates the DNA recombination and the initiation of the genomic alteration. An extensive collection of mice have been generated, each line expressing Cre from a promoter that is either tissue specific, cell specific, developmentally specific or responsive to an exogenous agent like tetracycline. With such a collection available, several promoter-specific mouse models can be studied in parallel. Additionally, researchers have generated an extensive collection of vectors that express Cre recombinase from a reliable promoter, and transient expression of Cre results in high rates of recombination in cultured cells. Thus, recombination can be triggered in ES cells to generate a more traditional knockout mouse in addition to the tissue-specific knockout. Alternatively, the mouse can be bred and grown as a pseudo- wild type with out any recombination, and then a population of cells cultured from this mouse can be transfected with a Cre-expression vector to generate recombined cells.
Recently, Flp recombinase (and its frt DNA sites) have also proven useful in mouse transgenics (9,10). Although few lines of mice have been generated to express Flp in vivo, this system is very useful for the removal of the selection gene from the targeted gene at the ES cell stage. The presence of a Neomycin resistance cassette in an intron can result in an alteration of gene function and therefore produce an unwanted or even lethal phenotype (11). This problem can be avoided if the investigator utilizes both the Cre and Flp recombination systems. A targeting vector containing both a Flp-flanked neoR marker and a loxP-flanked exon can be introduced into ES cells. After selection, the Neomyocin resistance cassette can be removed with Flp recombinase before the ES cells are injected into host blastocysts to make mice. (See Figure 4) With this system, the chimeric offspring contain only a minimal genetic modification (the addition of two loxP sites and one Frt site) in the gene of interest, limiting the likelihood of a complicating phenotype. As with a loxP-only targeting, the regulated expression of Cre results in the regulated alteration of this gene.
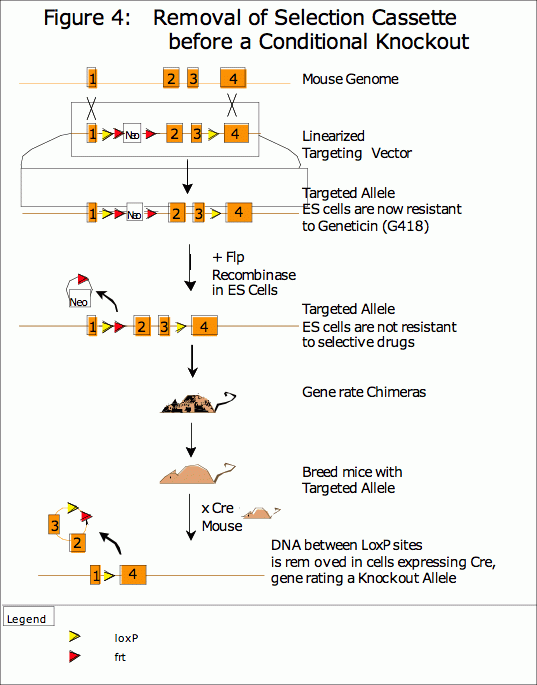
Glossary
Bacteriophage
A virus that infects bacteria
Cis orientation
Two LoxP sites are in CIS on a piece of DNA if they are in opposite directionalities. The sequences of CIS LoxP sites mirror one another.
Exogenous expression
Non-typical expression of a gene, usually due to a change in or replacement of the promoter of the gene. Can cause an expression level that is higher, lower or differently regulated for that cell type.
Exon
A portion of a gene that contains sequence that codes for the protein
Floxed
Flanked by loxP sites.
Gene Enhancer
A region of DNA that is separate from the Gene Promoter that also affects the transcription of the gene. Enhancers have been found within introns or even several kilobases from the 5' or 3' end of the gene.
Gene Promoter
A regulator region of DNA a short distance from the 5' end of a gene that acts as the binding site for RNA polymerase.
Gene trap
A sequence of DNA that is designed with at least (1) a splice acceptor to insert itself into genes and (2) a selection cassette to disrupt transcription. See The Center for Modeling Human Disease for a great explanation.
Intron
A non-coding sequence located between exons of a gene.
PCR
Polymerase chain reaction- a method for amplifying specific DNA segments which exploits certain features of DNA replication.
Southern Blot
Transfer of electrophorectically separated fragments of DNA from the gel to an absorbent sheet such as paper. This sheet is then immersed in a solution containing a labeled probe that will bind to a fragment of interest.
Trans orientation
LoxP sites are in TRANS if they all have the same directionality.
Vector
In cloning, the plasmid or phage chromosome used to carry the cloned DNA segment.
References
- Smithies, O. et al. (1985). Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 317:230-4.
- Sauer, B. and N. Henderson. (1998). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. PNAS USA. 85:5166-70.
- Vooijs, M et al. (1998). Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene. 17:1-12.
- Dymecki, S.M. and H. Tomasiewicz. (1998). Using Flp-recombinase to characterize expansion of Wnt1-expressing neural progenitors in the mouse. Dev Biol. 201:57-65.
- Scacheri, P.C. et al. (2001). Bidirectional transcriptional activity of PGK-neomycin and unexpected embryonic lethality in heterozygote chimeric knockout mice. Genesis. 30:259-63.