What is a nanowell device?
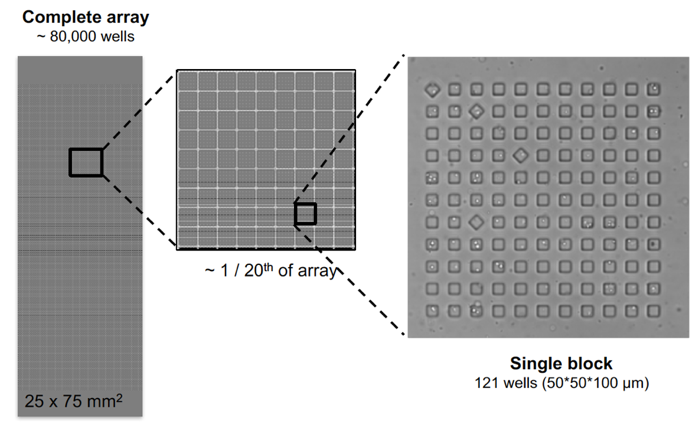
An array of wells, manufactured from PDMS (a silicone-based organic polymer), using photolithography and replica molding. The device is comprised of blocks of subnanoliter wells within a 1” X 3” glass slide. Each array has roughly 85,000 wells, which can hold 1 to 5 cells each.Nanowell Arrays are manufactured by the Nanowell Cytometry Platform for use in single cell phenotyping and SeqWell single cell RNAseq.
Cell Recovery
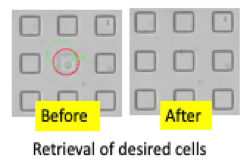
Recovery of single cells from arrays by alignment of microengraving and/or imaging data sets with the array grid, identification and location of individual cells, and picking/transfer for culture and expansion or genome or expression analysis.
SeqWell
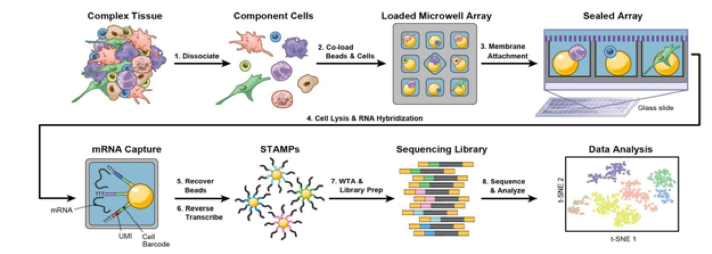
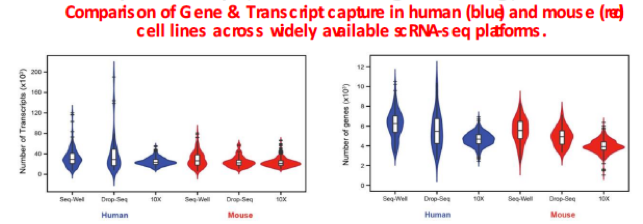
A relatively high throughput, cost-effective method for single cell RNAseq developed by Koch Institute investigators Chris Love and Alex Shalek. Individual cells are seeded into nanowell arrays and lysed, and the associated mRNA is attached to polyT containing beads. We work in collaboration with the Koch Institute Integrated Genomics & Bioinformatics Core/MIT BioMicro Center to support library preparation, quality control, sequencing and analysis.
For information on initial access, assisted service and training, please review our SeqWell Guidelines and these SeqWell Protocols.